Considerable effort and hope has been expressed in the literature for the potential benefit of exploiting the distinguishing MRI features of the diseased prostate. Some researchers used a single modality such as Apparent Diffusion Coefficient (ADC), Diffusion Weighted Images (DWI), Dynamic Contrast Enhancement (DCE), T2 to detect and localize the tumors within a prostate. In these cases, statistical averages of a given MRI modality over a given region of interest (ROI) delineated by the radiologist are computed and used as a metric of disease. The measurements were compared to the “gold standard” of assessment of histology slides taken from prostatectomy specimens processed in MRI-based molds. More commonly, other researchers have combined two or more of the modalities in the Multi-Parametric (MP) MRI approach to detect and localize the disease. The use of multiple sets of data tends to increase the sensitivity and specificity for finding the disease. The two or more sets of MRI are not spatially registered at the pixel level to each other. However, the multiple MRI images are correlated with each other, and the statistics of the region of interest (ROI) are separately determined by the researchers and compared to the pathologist’s evaluation of the histology. Increasing number of modalities generally elevate the sensitivity and specificity. Similarly, researchers used single modalities such as ADC, DWI, DCE, T2 to Gleason score and assess the disease. They compare the statistical metric with the histology assessment of the Gleason score or expected tumor aggressiveness. The use of multiple sets of data generally increases the sensitivity and specificity for scoring the disease. However, a severe limitation of these studies is the lack of a consistent, coherent approach that can be applied to all clinics and patients to support protocols, operate at the voxel level, is often only qualitative, and can depend on the observations of a trained radiologist.
This research effort analyzes the spectral distribution only and departs from more standard Computer Aided Diagnosis (CAD) algorithms that depend solely on spatial analysis of a specific MP MRI modality. In addition, this proposed research does not emulate radionomics that rely on extracting spatial features from a single image modality. Most approaches that discriminate between cancer and normal tissue or Gleason score depend on spatial processing of an image and that may assess textures such as the local roughness, smoothness etc. in order to distinguish cancer and normal tissues or evaluate the tumor aggressiveness. Texture-based imaging features in conjunction with machine learning-based classifications have been applied for classifying malignant from noncancerous prostate tissues. Texture and spatial processing requires setting fixed spatial windows to assess the local environment. Because these windows are fixed, the texture and spatial processing can miss detecting tumors that can vary considerably in size, especially missing small lesions.
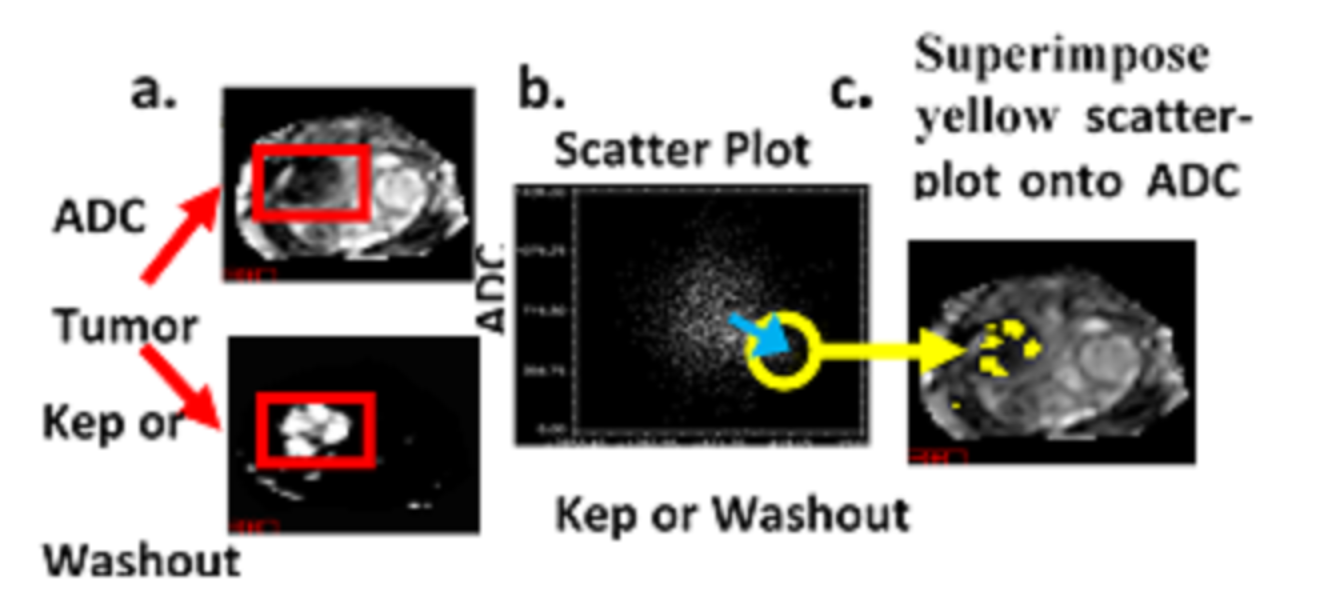
Current methods for assessing the location of prostate lesions divide the prostate into only 20 to 50 segments. These methods fail to fully exploit MRI’s high spatial resolution. Furthermore, these current techniques do not evaluate the variable aggressiveness inside the tumor and only summarize the lethality with a single metric, despite the considerable heterogeneity inside the tumor. In addition, the parameters used in the malignancy probability and Composite Biological Score (CBS) are fixed from patient to patient in order to determine whether a voxel is a lesion or normal tissue. These parameters may vary for each patient and these current techniques may not offer a robust solution for assessing patients.
Comparing the scoring results using image-based biomarkers and supervised target detection with other approaches is currently problematic. The embodiment offers the first description of this technique but in the future after further validation this technique will be compared with other approaches. For example, Pi-Rads v2 [41] is relatively new (2016) and requires experienced and specially trained radiologists to examine the entire prospective tumor, rather than evaluate every voxel within the tumor. Currently Pi-Rads v2 assessments are relatively rare compared to the more conventional histological determination of Gleason score tumor volume. The embodiment, however, employs relatively few radiologists. Other approaches use self-training and learning approaches [17, 19, 55] to detect tumors.